If you had asked me two weeks ago how to pronounce Anthoceros, I would have confidently (or, as confident as I ever am with these naming things) replied: AnthoCERos. Hard push on the ‘cer’.
But if Anthoceros means ‘flower horn’, and has the same etymological roots as the ‘nose horned’ Rhinoceros, well then, shouldn’t I be pronouncing it AnTHOceros?
While the recently published Anthoceros genome didn’t manage to answer this question for me, it did touch on some other cool facts about this long-neglected group of plant.
Before we begin, it’s important to clarify that despite ‘flower horn’ name, Anthoceros species aren’t angiosperms and therefore do not actually have flowers. In fact, the division Anthocerotophyta, commonly referred to as hornworts, are ‘simple’ plants. Which is a snobby way of saying that they reached land and then ‘took a bit of an evolutionary development break’, instead of going on to acquire things like vascular tissue, seeds, or flowers. As with other ‘simple’ plants (I’m looking at you moss and liverwort), hornworts are generally found hanging out in damp and humid patches of ground. Sometimes, rarely, they make it up onto the bark of trees.
Physically, hornworts also look a bit simple, consisting pretty largely of flat structures, of course with the exception of their horns. They also (again like mosses and liverworts), have less-than-the-usual amount of chromosomes: these guys spend their days hanging out as haploid gametophytes containing half of the normal set of chromosomes, in contrast with most of the other living things we see, which like to be diploid. At some point, the gametophyte plants create structures called sporophytes, dedicated to making reproductive spores. In the case of hornworts, those sporophytes just happen to look like little horns. Thus the name.
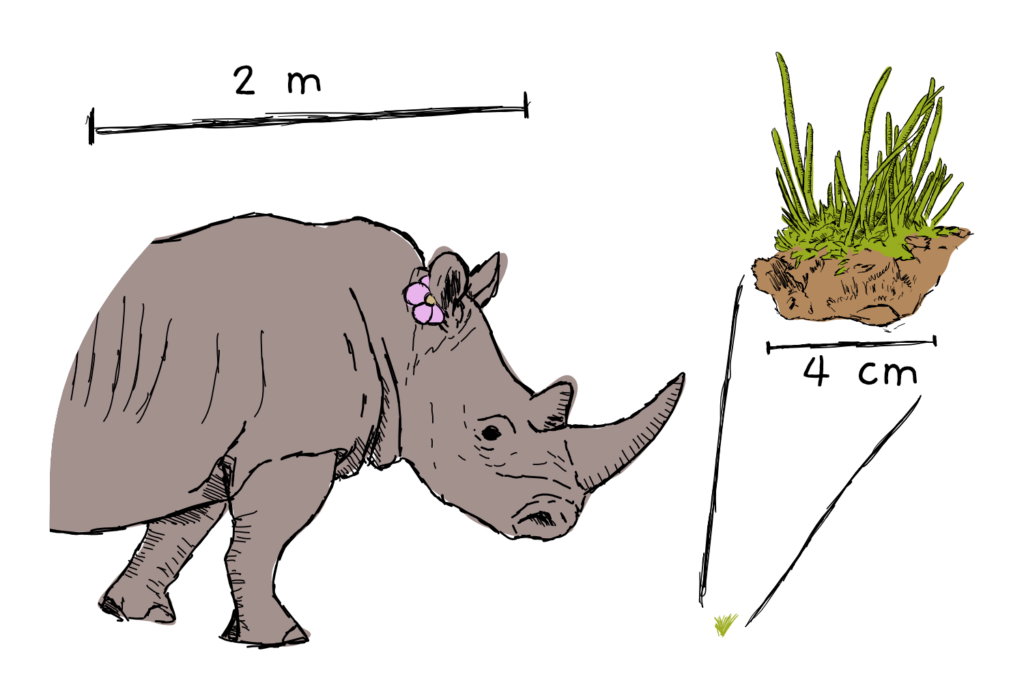
Here’s a picture of a hornwort next to a rhinoceros with a flower in his ear, just to make the whole thing super clear:
A bunch of evidence suggests that this ‘simple’ group of mosses, liverworts and hornworts- collectively referred to as ‘bryophytes’- represent ‘early’ land plants. Which means that understanding how these plants survive might help us understand the features that allowed plants to first colonise, and then dominate this now-green earth of ours!
Of the three bryophyte bunches, hornworts were the only group that didn’t have a member with a sequenced genomes. Possibly because, in the past, there has been some confusion about whether they are their own separate taxonomical group (they are), or just a member of the moss gang. But also perhaps because hornworts contain way fewer species than most other plant lineages- only about 200-250 species in total. But despite their small numbers, hornworts can still do some cool things that the others don’t. For example, hornworts form tight symbiotic relationships with many cyanobacteria and fungi, and develop special ‘mucilage-filled cavities (yep, gross eh?) to house these co-workers in. Even more stunningly, they have chloroplasts that contain pyrenoids- specialised carbon concentrating compartments that are found in algae, but don’t exist in any other land plant.
(Carbon concentration is a pretty hot topic when discussing how to improve photosynthesis. We’ve mentioned it briefly before over here and here.)

Thanks to Jian Zhang and colleagues, we now have the first draft genome of a hornwort species: Anthoceros angustus. The group used a dual-sequencing method, complementing Illumina sequencing, which gives highly accurate information in small snippets of just a couple of hundred base pairs, with Oxfort Nanopore sequencing, which gives long stretches of sequence that tend to have quite a few mistakes. The Nanopore provides the physical structure of the genome, while the Illumina gives it accuracy. After a whole lot of processing, including filtering, error-correction and decontamination (removing sequences that probably belong to some of those symbionts we mentioned), the group had a 119 Mb genome in front of them.
Once they had the genome, they had to work out what was actually in it. As with most species, a large chunk of the sequences (64% of the assembled genomes) were repeats- mostly consisting of transposable elements. But of course, there was also a whole lot of protein coding genes (mRNA- 14,629 predicted), tRNA (347) and ribosomal RNAs (94). Th authors also discovered a bunch small- or micro- RNAs, species that generally have a role in regulating gene expression, including some miRNA conserved in angiosperms.
In order to see if the A. angustus genome was hiding anything special, the scientists compared the proteins predicted from their new genome with that of 18 other green species, including 11 land plants, and seven algae (two charophytes, five chlorophytes). Most of the gene families that the hornwort shared with its green friends looked more similar to things found in land plants, as opposed to algae, and only a small number (about 1%) looked a lot like those found in algae. (Although hornworts are the only land plants to have functional pyrenoids, many of the components needed to make the pyrenoids are also found in non-angiosperm land plants. These species have just managed to ‘lose’ important parts of the machinery and can no longer make the complete thing). A. angustus also had 497 gene families that look to be unique, which cover a range of possible functions.
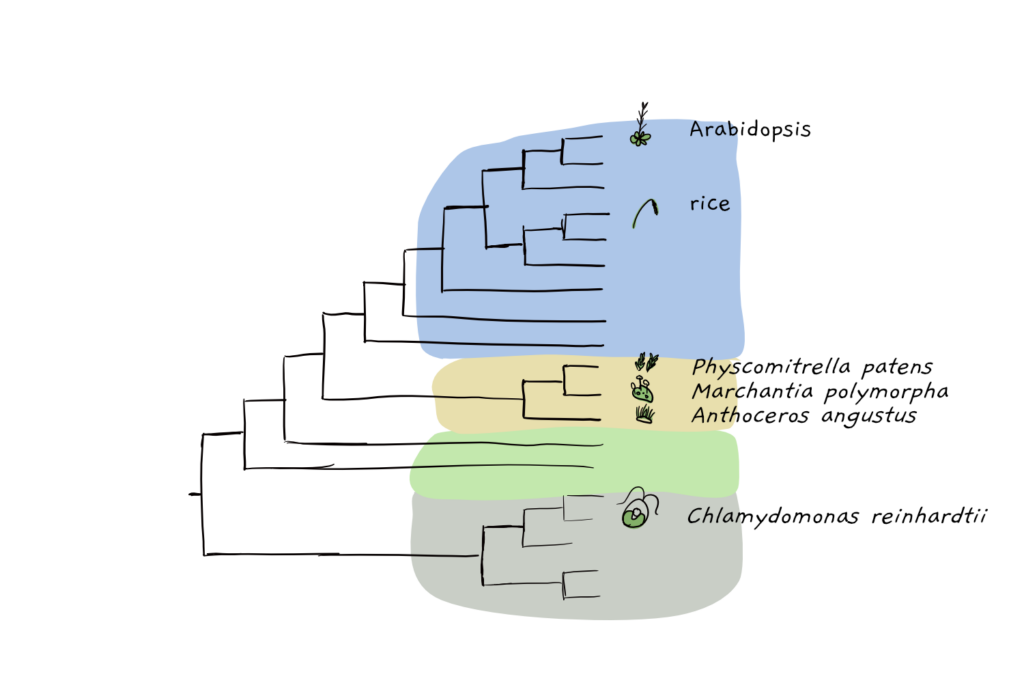
As expected from previous studies, a phylogentic tree, indicating the evolutionary relationship of different green species, showed clustering of A. angustus with the other bryophyte species Physcomitrella patens (moss) and Marchantia polymorpha (liverwort). The hornworts probably split off about 275 million years ago during the middle Carboniferous to early Jurassic period, aka, before Stegosaurus and Allosaurus had even thought to walk the lands. So far, the earliest fossil record we have of hornworts, a hornwort spore from Argentina, dates back to only about 145 million years ago.
In those few hundred million years that hornworts have been hanging around the corners of the earth, they’ve managed to gain a few gene families, and have lost a whole lot more. Most land plants, including moss, have copy-pasted their entire genome one or more times, an easy way to immediately create extra genes which can then evolve new and specialised functions. But hornworts, like their liverwort sisters, have avoided this process- known as whole genome duplication- instead opting for a more streamlined genome. Perhaps because of this, hornwort has a fairly reduced set of transcription factors, which regulate when and where genes are expressed. Of course, they also don’t have much need for genes involved in regulating the more complex embryogenesis processes found in seed plants, nor do they need those genes that dictate more complex body plans. Interestingly, some genes that we know to have function in seeds, are found in the hornwort, although with different uses. ABI3 genes, for example, are linked to seed desiccation tolerance in flowering plants. In bryophytes, the exist, but instead play a role in desiccation tolerance of the vegetative tissue.
What hornworts do have a whole lot of, is PPR proteins. PPRs (pentatrico-peptide repeat proteins), are found across the plant kingdom, and are involved in processing of organellar RNA. A massive 7.9% of the hornwort genome is PPRs! Among other things, PPRs are involved in RNA editing, a process in which mRNA ‘C’ bases are changed to ‘U’s. It’s been proposed that PPRs can therefore help ‘fix’ the genome mistakes that have arisen by DNA damage. This feature isn’t present in algae, leading to the suggestion that moving away from a UV-protecting water environment might have necessitated this last-minute repair process. Similarly, the A. angustus has an expanded amount of genes involved in the synthesis of UV-B protecting flavenoids, and cupins and abscisic acid catabolism genes, both linked to drought stress tolerance.
Another way of dealing with stress, apart from making your own new genes, is to simply steal some genes! Like its moss and liverwort siblings, hornwort was found to contain a bunch of genes (19), stolen from a whole lot of different bacterial and fungal species (14 families!). It’s possible that at least some of these genes were taken from hornwort symbionts. Generally, the genes seem to help the hornwort do things like survive stress, be more metabolically flexible in changing environments, and defend itself against herbivores or pathogens.
All together, the A. angustus genome is an important piece of our plant-understanding puzzle, while also helping to provide specific clues as to how plants invaded and then proliferated on land.
I wanted to end this post by childishly pointing out that, while Anthoceros has a published genome, Rhinoceros does not. But it turns out that may not be true. So I’m just going to stick with out standard insult: ‘way to not even fix inorganic carbon rhinoceros!’.
If you like reading about hornworts, you might also like to hear about their sister group- liverworts.
References:
OPEN ACCESS The hornwort genome and early land plant evolution. Jian Zhang, Xin-Xing Fu, Rui-Qi Li, Xiang Zhao, Yang Liu, Ming-He Li, Arthur Zwaenepoel, Hong Ma, Bernard Goffinet, Yan-Long Guan, Jia-Yu Xue, Yi-Ying Liao, Qing-Feng Wang, Qing-Hua Wang, Jie-Yu Wang, Guo-Qiang Zhang, Zhi-Wen Wang, Yu Jia, Mei-Zhi Wang, Shan-Shan Dong, Jian-Fen Yang, Yuan-Nian Jiao, Ya-Long Guo, Hong-Zhi Kong, An-Ming Lu, Huan-Ming Yang, Shou-Zhou Zhang, Yves Van de Peer, Zhong-Jian Liu & Zhi-Duan Chen. 2020. Nature Plants.
You should also go and check out the community post written by author Zhong-Jian Liu and Di-Yang Zhang over here.
Big shout out to T D S V for discussing Anthoceros (and the concept of ‘anthoceros green’) with me!